| Journal of Medical Cases, ISSN 1923-4155 print, 1923-4163 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Med Cases and Elmer Press Inc |
| Journal website https://www.journalmc.org |
Case Report
Volume 15, Number 9, September 2024, pages 250-255
Clinical, Phenotypic and Molecular Characterization of NUP214-ABL1 Fusion Positive Myeloid Malignancies
Chelby Wakefielda, c, Mary Hansen Smithb, Ulyana Dashkevycha, Maria Proytchevab, Sharad Khuranaa
aDepartment of Hematology and Oncology, University of Arizona, Tucson, AZ, USA
bDepartment of Pathology, University of Arizona, Tucson, AZ, USA
cCorresponding Author: Chelby Wakefield, Department of Hematology and Oncology, University of Arizona, Tucson, AZ, USA
Manuscript submitted July 2, 2024, accepted August 7, 2024, published online August 22, 2024
Short title: NUP214-ABL1 Fusion in Myeloid Malignancies
doi: https://doi.org/10.14740/jmc4286
| Abstract | ▴Top |
The identification of a NUP214-ABL1 fusion has been seen in about 6% of patients with T lymphoblastic leukemia (T-ALL). It has been described at a lower frequency in B-lymphoblastic leukemia (B-ALL) patients as well. To our knowledge, this is the first case report documenting a NUP214-ABL1 fusion in a patient with newly diagnosed myelodysplastic syndrome (MDS) as identified by next-generation sequencing (NGS). A case report by Wang et al recently described a case report of the first NUP214-ABL1 fusion in a patient with newly diagnosed acute myeloid leukemia (AML). This shows that this specific translocation is not isolated to lymphoid malignancies, and can be associated with myeloid malignancies as well. The potential use of tyrosine kinase inhibitors (TKIs) as a line of treatment for patients who harbor this translocation makes this finding of particular interest. However, while there have been individual reports noting the effect of TKIs in T-ALLs with NUP214-ABL1 fusions, additional research is needed to fully understand the role of this mutation in myeloid derived malignancies, and its corresponding treatment and prognostic implications.
Keywords: Myelodysplastic syndrome; Molecular characterization; Myeloid; Tyrosine kinase inhibitor
| Introduction | ▴Top |
Myeloid malignancies are clonal disorders of progenitor cells that can be broken down into categories that include myeloproliferative neoplasms (MPNs), myelodysplastic syndrome (MDS), MDS/MPN overlap diseases like chronic myelomonocytic leukemia (CMML), and acute phases such as acute myelogenous leukemia (AML) [1]. Approximately one-third of MDS transforms to AML over time; however, the risk varies depending on the MDS subtype including blast percentage and cytogenetic abnormalities. Risk stratification of an untreated MDS patient is crucial in clinical practice to help guide treatment choices [1-4]. The International Prognostic Scoring System (IPSS) is a tool which includes the blast percentage in the marrow, the type of somatic mutations identified in the marrow, and the number of cytopenias present at the time of bone marrow biopsy [2]. The Revised International Prognostic Scoring System (IPSS-R) was a modification to the IPSS made in 2012 [3] and as technology in medicine has advanced, the Molecular International Prognostic Scoring System (IPSS-M) has been developed, which includes the mutational status of 31 genes in addition to the factors included in the IPSS-R and IPSS [4].
Over the past decade, the understanding of genetic mutations in myeloid neoplasms including MDS has dramatically improved. The invention of next-generation sequencing (NGS) technology has allowed for the creation of large databases that have identified recurrent driver mutations in these disorders [5]. The number of mutations is known to vary depending on the disease subtype, with a median number of 6 per exome in low-risk MDS, 9 in MDS with excess blasts, 12 in CMML, 8 in MDS/MPN, and 13 in secondary AML. At this time, greater than 30 driver genes that are involved in the pathogenesis of MDS have been identified, and are categorized into several discrete functional pathways [5]. The most frequently observed are genes affecting epigenetic regulation, RNA splicing, DNA methylation, and chromatin/histone regulation. These are commonly, but not limited to: TET2, DNMT3A, IDH1/IDH2, MLL2, EZH2, ASXL1, ARID2, SF3B1, SRSF2, U2AF1, U2AF2. Other molecular mutations involving p53 regulation, cohesion proteins, transcription factors, the RAS pathway and other signaling molecules are commonly identified in MDS, and are well described in the literature. These include, but are not limited to: STAG2, RAD21, SMC1A, RUNX1, ETV6, GATA32, IRF1, NRAS, KRAS, NF1, JAK2, KIT, MPL, GNB1, FLT3. These molecular alterations allow excellent opportunity for targeted therapy, which has been effective in disorders such as AML and chronic myeloid leukemia (CML) for example, but has seen less application in the treatment of MDS. With added investigation, novel markers can be identified and therapeutic targets can be explored [5].
NUP214 is a component of the nuclear pore complex within a cell and plays a key role in protein and mRNA nuclear export. Chromosomal translocations that include this locus can be found in acute leukemias. Most notably, SET-NUP214 is most associated with acute lymphoblastic leukemia (ALL), while DEK-NUP214 is seen almost exclusively in AML. Both DEK and SET are chromatin remodeling proteins that play a role in transcription regulation and thus the fusion of them to NUP214 and their respective resultant leukemias point to different leukemogenic driver mechanisms [6]. The ABL1 locus is commonly associated with CML where it is fused with BCR, which has shown remarkable responsiveness to tyrosine kinase inhibition. NUP214-ABL1 fusion is less common and has been detected in 6% of T-ALL, and, less commonly, has been reported in B-ALL [7, 8]. As a constitutively active tyrosine kinase, NUP214-ABL1 may be an ideal target for tyrosine kinase inhibitor (TKI) therapy in a similar fashion as BCR-ABL1 fusion has been [9, 10]. However, to date, the therapeutic effect and utility of this treatment method for NUP214-ABL1 have not been fully explored and additional research is needed [11-16]. To our knowledge, there has been one case report documenting NUP214-ABL1 fusion in AML [17]. This supports the idea that this molecular rearrangement does occur in myeloid malignancies, not just lymphoid. Furthermore, the presence of a NUP214-ABL1 fusion gene in a patient with a myeloid malignancy has unclear significance at this time and requires additional research efforts to fully understand its role in this malignancy.
| Case Report | ▴Top |
The case was a 44-year-old female with a pertinent past medical history of viral encephalitis in childhood with subsequent intractable epilepsy and intellectual disability, who presented to an outside hospital in September of 2023 after having a seizure at home. She did not have any previous oncologic history or exposure to chemotherapeutics or radiation therapy. Upon evaluation at the outside facility, lab work revealed a hemoglobin (Hgb) of 3.6 g/dL, white blood cell (WBC) count of 1,500/µL, and platelet (plt) count of 21,000/µL. She was subsequently transferred to our facility for further workup of pancytopenia and concern for underlying hematologic malignancy.
Upon arrival at our facility, due to traumatic injuries from her seizure, a computed tomography (CT) of chest, abdomen and pelvis was performed which showed a large left-sided subcapsular hematoma measuring up to 3.5 cm encompassing greater than 50% of the surface of the kidney with resultant mass effect on the left renal parenchyma. Hgb was 8.1 g/dL following packed red blood cell transfusion prior to transfer, WBC count was 1,500/µL, plt count was 21,000/µL, fibrinogen was 19, and international normalized ratio (INR) was 1.2. Differential showed absolute neutrophilia (740/µL), absolute lymphopenia (680/µL), and absolute monocytopenia (20/µL). There were also a small number of intermediate to large blasts with fine chromatin, nucleoli and scant cytoplasm. Additionally, rare hyposegmented hypogranular neutrophils were also noted. Platelets were decreased with rare large agranular forms.
A bone marrow aspirate and biopsy was performed; however, the aspirate smears were paucicellular precluding differential count. Immunohistochemistry with CD34 highlighted about 15% blasts (Fig. 1). Concurrent bone marrow flow cytometry identified an abnormal CD34+ myeloid blast population representing 13% of the WBCs. The neoplastic cells were positive for CD11c, CD11b, CD13, CD33 (dim), CD117, myeloperoxidase (MPO) and human leukocyte antigen (HLA)-DR. They were negative for all monocytic, B- and T-cell markers included in the panel (Fig. 2). Peripheral blood flow cytometry also showed 14% CD34+ myeloid blasts. AML fluorescence in situ hybridization (FISH) testing identified a deletion 5q, gain of 5p, trisomy 8, gain of 11q23, and three copies of RUNX1 (21q22). NGS identified a NUP214-ABL1 fusion (locus chr9:134090755-chr9:133730188) (Fig. 3). Chromosome analysis showed abnormal female karyotype: 48,XX,3,+5,der(5)t(3;5)(q21;q15)x2,+8,+19,der(19)t(11;19)(q13;p13.3),idic(21)(p11.2)[cp12]/47,idem,t(4;13)(q31.3;q12),-der(19),-idic(21),+21[6]/4748,idem,add(18)(p11.2)[cp2]/46,XX[4] (Fig. 4).
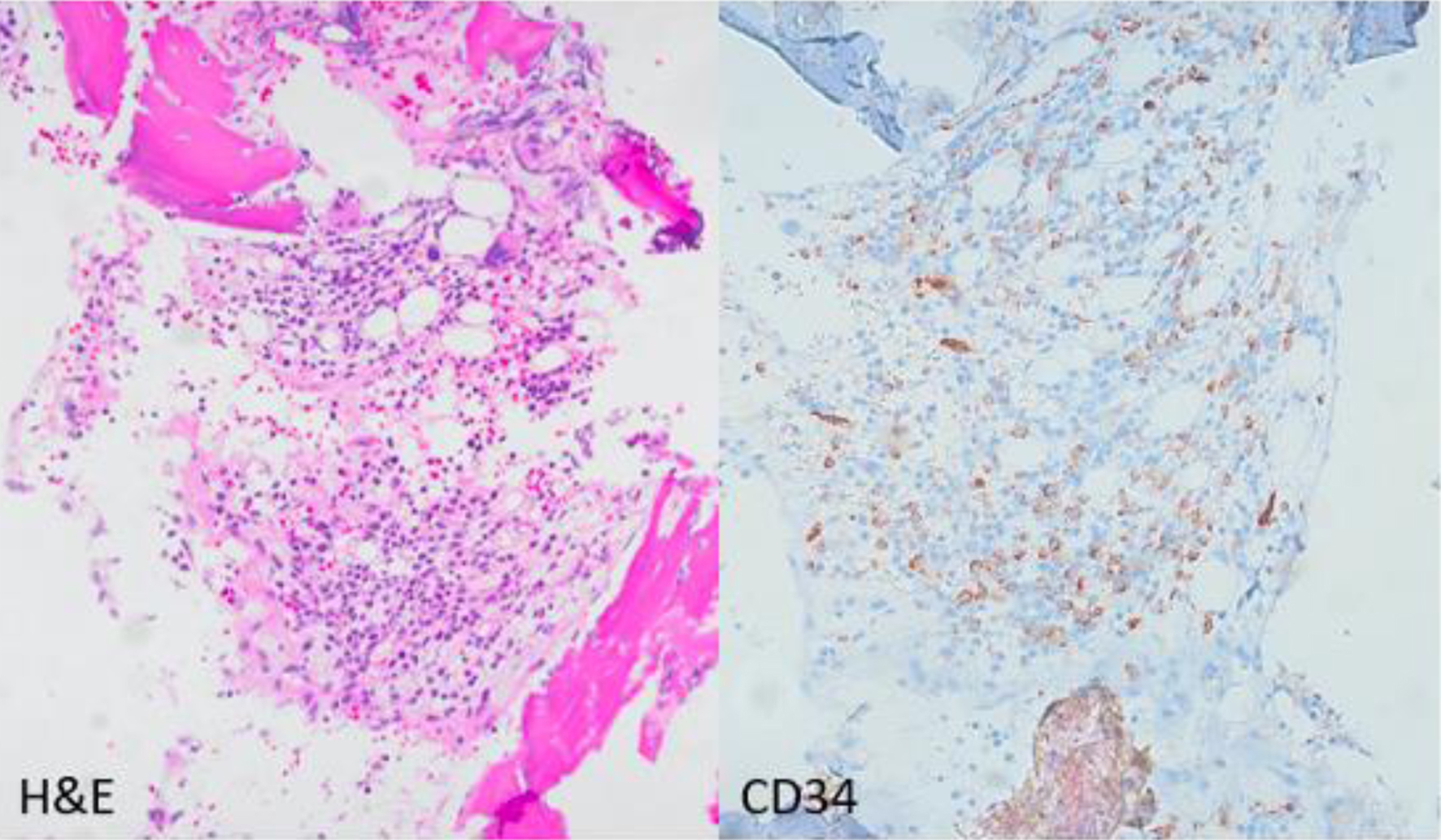 Click for large image | Figure 1. Photomicrograph (× 200) of the bone marrow core biopsy highlighting CD34 positive blasts comprising approximately 15% of the bone marrow elements. Hematoxylin and eosin stain (H&E) on left, CD34 immunohistochemical stain on the right. |
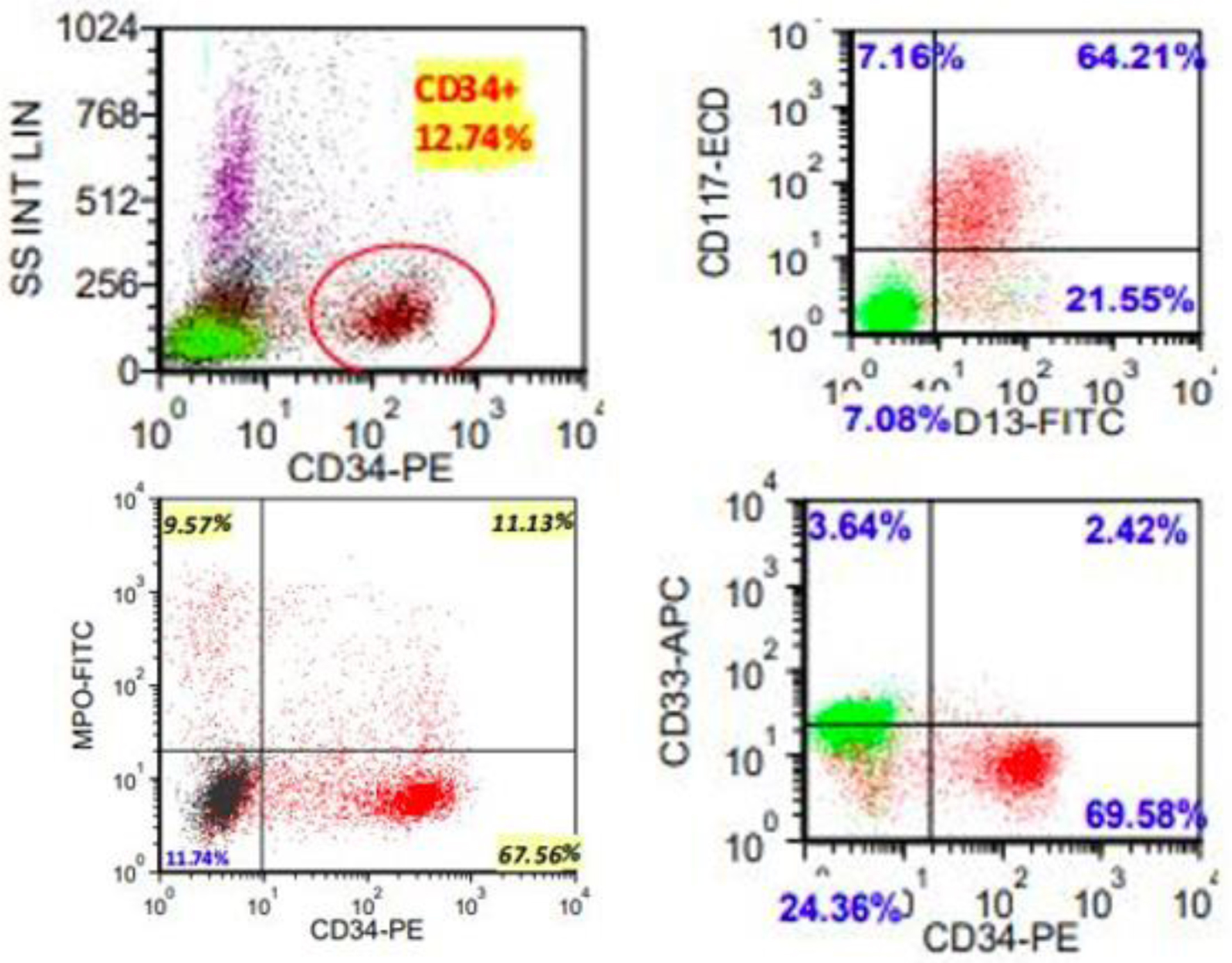 Click for large image | Figure 2. Representative flow cytometric scatter plots highlighting myeloblast population. Full blast immunophenotype: positive for CD10 (subset), CD11b (subset), CD11c, CD13, CD33 (dim), CD117, MPO (small subset), and HLA-DR (dim); negative for TdT, CD2, sCD3, cCD3, CD5, CD4, CD8, CD19, CD22, CD25, CD64 and CD123. |
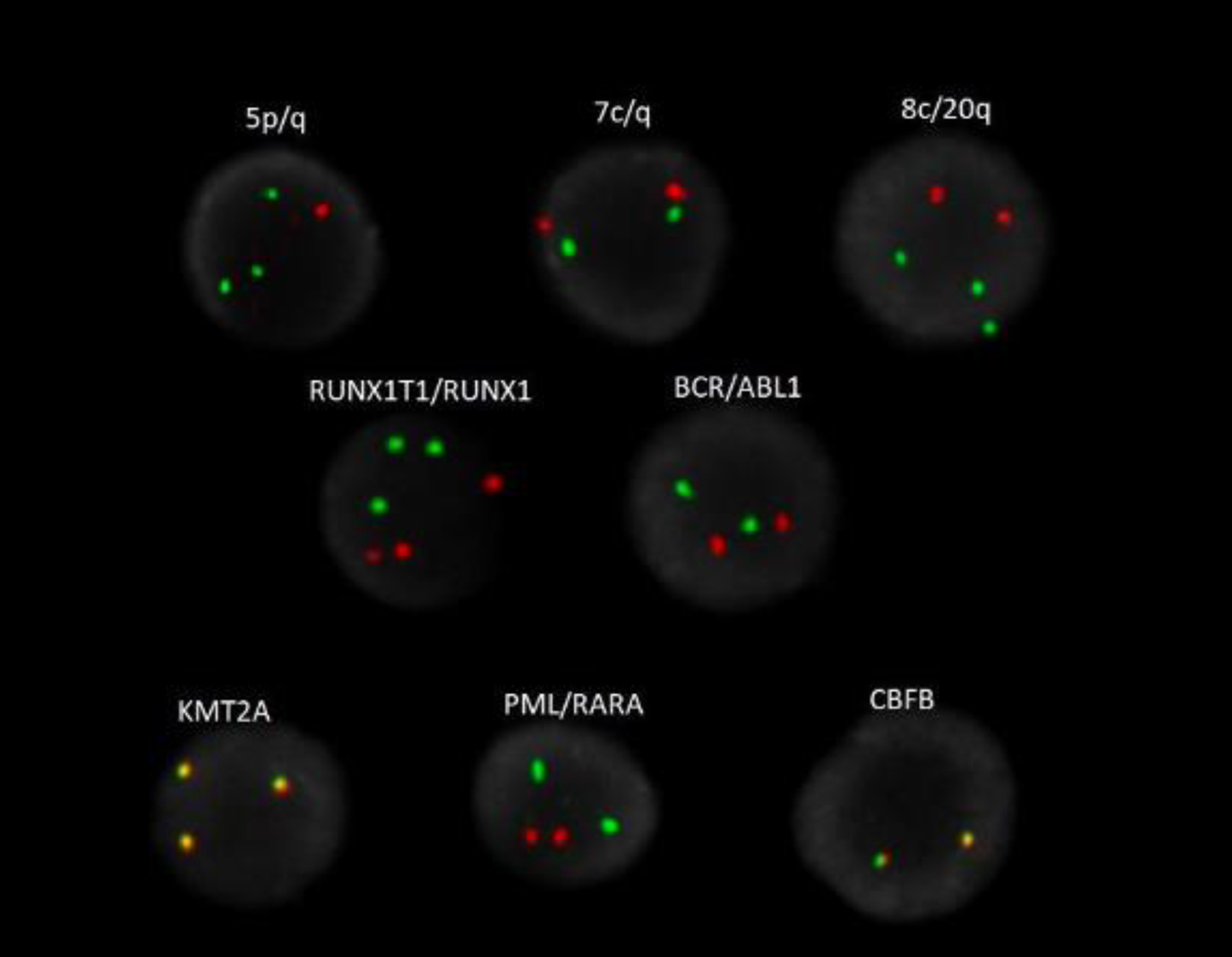 Click for large image | Figure 3. AML FISH analysis highlighting 5q deletion (red), gains of 5p (green), trisomy 8 (green), gains of 11q23 (KMT2A), three copies of 8q22 (RUNX1T1, orange) and 21q22 (RUNX1, green). |
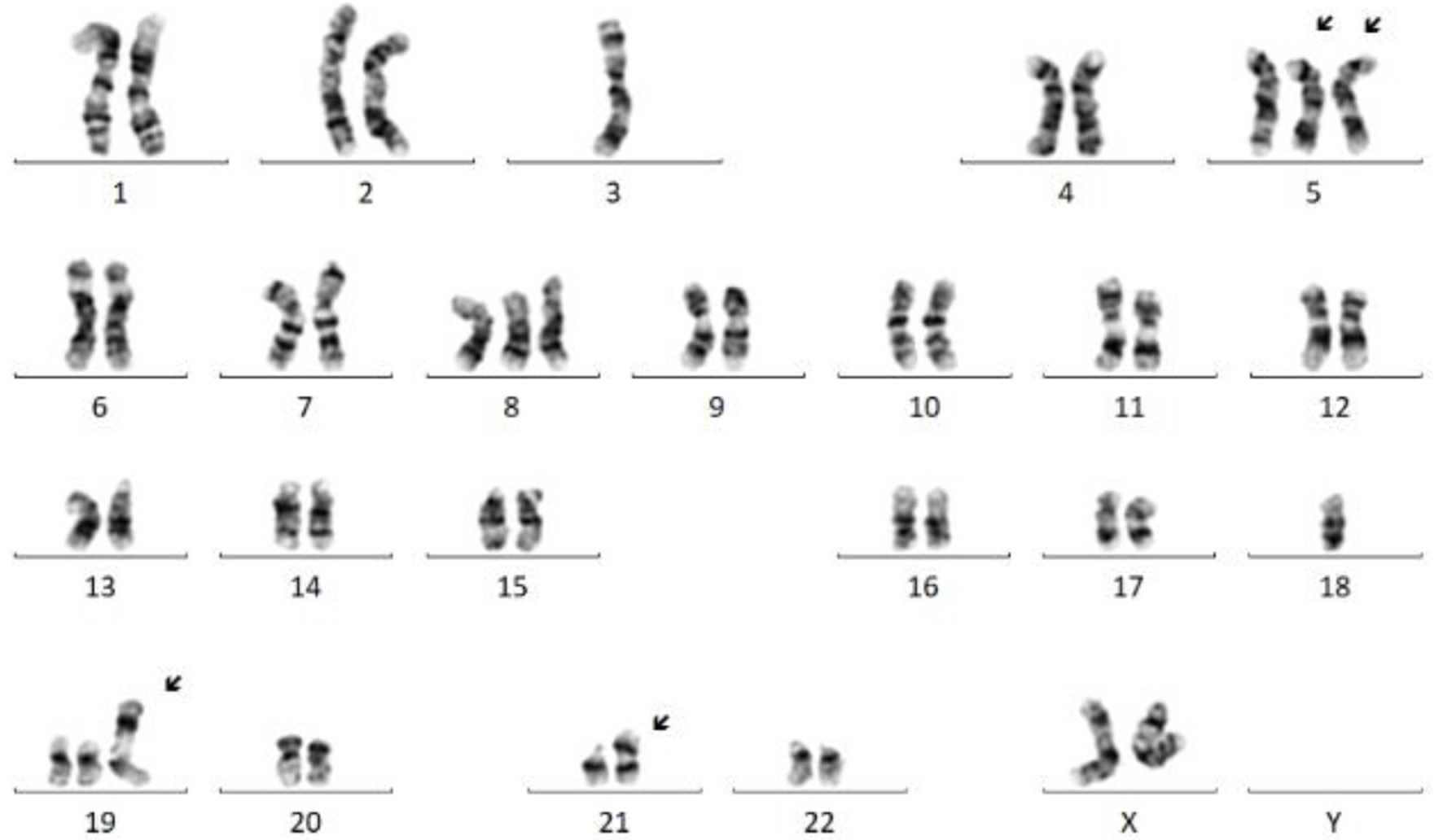 Click for large image | Figure 4. Representative karyogram exhibiting complex female karyotype with an unbalanced translocation between 3q and 5q resulting in deletions of 3p and 5q; trisomies 5 (partial, leading to gain of 5p), 8, and 19; an isodicentric chromosome 21 leading to one extra copy of 21q/RUNX1; and another unbalanced translocation between 11q and 19p leading to gain of 11q/KMT2A. Multiple clones were present, exhibiting similar abnormalities with minor differences. |
Based on these findings, the final diagnosis was considered to be MDS with excess blasts (MDS-EB2) according to the International Consensus Classification of Myeloid Neoplasms, 2022 [1]. Prognosis, utilizing the IPSS-R and IPSS-M prognostic risk assessment for MDS, was 9.50 and 3.79, respectively, with a score of 9.44 for IPSS-R age-adjusted score. This conferred a very poor and adverse prognosis respectively.
Our patient did not have decision making capacity, which was long-standing, and her health care decisions were made by her mother who held medical power of attorney. After extensive conversations regarding prognosis and what the treatment landscape would entail, it was ultimately decided by the patient’s family to forego treatment and pursue a hospice- and comfort-centered treatment approach.
| Discussion | ▴Top |
This study documents, as far as we know, the first report of a patient with MDS-EB2 and a NUP214-ABL1 fusion on molecular analysis. The overall impact of this chromosomal abnormality in this disease entity is yet to be fully understood. The treatment outcomes over the past 30 years in AML and MDS have improved significantly. This is largely due to the advancement of our identification strategies including FISH and molecular testing, which allows for a superior understanding of the driving forces of cancer. Furthermore, with this advanced understanding, it is possible to define molecular targets and enhance treatment strategies to target them, as seen with FLT3 and IDH1/2 mutations in AML for example. In CML, ABL1 has fused with BCR, and with the discovery of TKIs, that fusion can be targeted and durable remissions can be achieved. Limited reports have documented the use of TKIs in targeting the novel NUP214-ABL1 fusion in T-ALL with interesting results; however, additional research is needed to better elucidate if TKIs have a substantial impact on this mutational prospect in myeloid derived malignancies.
One example is a case report by Nardi et al [18] who identified a NUP214-ABL1 fusion by multiplexed targeted RNA sequencing, at the time of relapse in a patient with Ph-negative B-ALL. The fusion escaped identification at the time of diagnosis and was noted to be present after receiving multi-chemotherapy induction and inotuzumab immunomodulatory therapy. Dasatinib was initiated in combination with inotuzumab, resulting in a complete remission (CR). However rapid relapse occurred with dasatinib alone. Deep remission was recaptured after he was transitioned to blinatumomab and ponatinib in advance of allogeneic hematopoietic stem cell transplantation. This case supports the idea that inventive targeted strategies need to be investigated and explored to bring meaningful results to difficult-to-treat disease entities and highlights the successes that they can achieve.
While the above case demonstrates NUP214-ABL1 as a potential novel therapeutic target in ALL, Wang et al [17] showed that this specific translocation is not isolated to lymphoid malignancies. Their case involves a male patient of 42 years of age who had presented to their facility with skin bleeding. WBC count was elevated at 82,500/µL (differential: neutrophils 2.7%, eosinophils 0.5%, basophils 0.1%, lymphocytes 13.7%, monocytes 83%), Hgb was 9.2 g/dL, and platelets count was 10,000/µL. Bone marrow aspiration revealed 78% blast cells which were positive for cytoplasmic MPO, CD7, CD13, CD33, CD34, CD38 (dim), CD56 (partial), CD117, CD123, and HLA-DR, and negative for cytoplasmic CD3, cytoplasmic CD79a, CD1a, CD3, CD4, CD5, CD8, CD10, CD11b, CD14, CD15, CD16, CD19 and CD64 by flow cytometry confirming a diagnosis of AML. NGS revealed a CEBPA (NM_004364.3) double mutation (p.K304_Q305insL, and p.D75Gfs*33), NRAS (NM_002524.5) point mutation (p.G13D), and a NUP214 (NM_005085.4)-ABL1 (NM_007313.2) fusion gene (fusion of NUP214 exon 34 and ABL1 exon 3). No amplification of the ABL1 gene was seen by interphase FISH. He was induced with idarubicin and cytarabine, achieving a morphological CR with minimal residual disease under 0.01%. Consolidation with Hi-DAC was started sequentially and as of his third cycle, he reportedly remained in CR. This is reportedly the first documented case of a NUP214-ABL1 fusion gene in a patient with AML in the literature.
NUP214-ABL1 is a translocation that is uncommon in hematologic malignancies; however, when it occurs it is often observed in lymphoid malignancies. As previously described, Wang et al identified this mutation for the first time in a patient with AML in 2021 [17]. We subsequently identified this mutation in a patient with MDS, which suggests this is a mutational anomaly not exclusive to lymphoid malignancies. Similar ABL1 translocations have been well characterized in other malignancies of myeloid origin such as CML, and these mutations can be targeted with TKIs that have the ability to control the disease for many years. It is unclear if these TKIs have meaningful activity in the treatment of NUP214-ABL1 translocations, both in lymphoid and myeloid malignancies. While additional information is still needed regarding the pathobiology and the significance of a NUP214-ABL1 fusion in a patient with a myeloid malignancy, the identification of a potential new molecular target and/or prognostic indicator is promising in these devastating malignancies.
Learning points
This case aims to highlight the significance of novel molecular markers that are commonly found during routine evaluation and workup of malignancy, in this case, a NUP214-ABL1 translocation. This specific mutation has been identified, albeit rarely, in lymphoid malignancies; however, this is the first case that it has been identified in a patient with MDS. These malignancies are driven by mutational abnormalities, many of which we are just starting to understand. The importance of identifying and further understanding these mutations can lead to potential treatment development.
Acknowledgments
None to declare.
Financial Disclosure
None to declare.
Conflict of Interest
None to declare.
Informed Consent
Informed consent has been obtained.
Author Contributions
C Wakefield: concept development, manuscript creation, and submission of manuscript; M Hansen Smith: procurement of images and figures, and generation of captions for images/figures; U Dashkevych: direct clinical care of the patient; M Proytcheva: procurement of images and figures, and manuscript oversight and editing; S Khurana: concept development, manuscript oversight and editing, and direct clinical care of patient.
Data Availability
The authors declare that data supporting the findings of this study are available within the article.
| References | ▴Top |
- Garcia-Manero G. Myelodysplastic syndromes: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol. 2023;98(8):1307-1325.
doi pubmed - Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, Sanz M, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079-2088.
pubmed - Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, Bennett JM, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454-2465.
doi pubmed pmc - Bernard E, Tuechler H, Greenberg PL, Hasserjian RP, Arango Ossa JE, Nannya Y, Devlin SM, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid. 2022;1(7):EVIDoa2200008.
doi pubmed - Ogawa S. Genetics of MDS. Blood. 2019;133(10):1049-1059.
doi pubmed pmc - Mendes A, Fahrenkrog B. NUP214 in leukemia: It's more than transport. Cells. 2019;8(1):76.
doi pubmed pmc - Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, Vermeesch JR, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36(10):1084-1089.
doi pubmed - Tsujimoto SI, Nakano Y, Osumi T, Okada K, Ouchi-Uchiyama M, Kataoka K, Fujii Y, et al. A cryptic NUP214-ABL1 fusion in B-cell precursor acute lymphoblastic leukemia. J Pediatr Hematol Oncol. 2018;40(6):e397-e399.
doi pubmed - De Keersmaecker K, Rocnik JL, Bernad R, Lee BH, Leeman D, Gielen O, Verachtert H, et al. Kinase activation and transformation by NUP214-ABL1 is dependent on the context of the nuclear pore. Mol Cell. 2008;31(1):134-142.
doi pubmed - Graux C, Stevens-Kroef M, Lafage M, Dastugue N, Harrison CJ, Mugneret F, Bahloula K, et al. Heterogeneous patterns of amplification of the NUP214-ABL1 fusion gene in T-cell acute lymphoblastic leukemia. Leukemia. 2009;23(1):125-133.
doi pubmed - Deenik W, Beverloo HB, van der Poel-van de Luytgaarde SC, Wattel MM, van Esser JW, Valk PJ, Cornelissen JJ. Rapid complete cytogenetic remission after upfront dasatinib monotherapy in a patient with a NUP214-ABL1-positive T-cell acute lymphoblastic leukemia. Leukemia. 2009;23(3):627-629.
doi pubmed - Stergianou K, Fox C, Russell NH. Fusion of NUP214 to ABL1 on amplified episomes in T-ALL—implications for treatment. Leukemia. 2005;19(9):1680-1681.
doi pubmed - Koschmieder S, Burmeister T, Bruggemann M, Berkemeier A, Volpert S, Wieacker P, Silling G, et al. Molecular monitoring in NUP214-ABL-positive T-acute lymphoblastic leukemia reveals clonal diversity and helps to guide targeted therapy. Leukemia. 2014;28(2):419-422.
doi pubmed - Tsurusaki Y, Nagai JI, Fujita S, Sugiyama M, Nakamura W, Hayashi A, Miyagawa N, et al. Whole-exome sequencing reveals the subclonal expression of NUP214-ABL1 fusion gene in T-cell acute lymphoblastic leukemia. Pediatr Blood Cancer. 2020;67(1):e28019.
doi pubmed - Aldoss I, Pullarkat V. Response to single agent dasatinib post allogeneic transplant in B-cell acute lymphoblastic leukemia with NUP214-ABL1. Leuk Lymphoma. 2019;60(11):2832-2834.
doi pubmed - Chen Y, Zhang L, Huang J, Hong X, Zhao J, Wang Z, Zhang K. Dasatinib and chemotherapy in a patient with early T-cell precursor acute lymphoblastic leukemia and NUP214-ABL1 fusion: a case report. Exp Ther Med. 2017;14(5):3979-3984.
doi pubmed pmc - Wang HP, He JJ, Zhu QY, Wang L, Li JH, Huang JS, Xie WZ, et al. Case report: the first report of NUP214-ABL1 fusion gene in acute myeloid leukemia patient detected by next-generation sequencing. Front Oncol. 2021;11:706798.
doi pubmed pmc - Nardi V, McAfee SL, Dal Cin P, Tsai HK, Amrein PC, Hobbs GS, Brunner AM, et al. Chemotherapy resistance in B-ALL with cryptic NUP214-ABL1 is amenable to kinase inhibition and immunotherapy. Oncologist. 2022;27(2):82-86.
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Medical Cases is published by Elmer Press Inc.


